Abstract
Background
Beyond carrying the plant embryo, seeds harbour intricate microbial communities whose transmission across successive plant generations can significantly influence the ecological and evolutionary dynamics of plant–microbe symbioses. The process of plant domestication has potential repercussions in genes involved in plant-microbiome interactions. However, the extent to which breeding can impact the seed microbiome is sparsely explored. Cannabis is a high-value crop but sparsely subjected to agricultural innovations established in other crop species during the last century. Here, we conduct a large-scale analysis of the bacterial seed microbiome of Cannabis across different domestication grades and investigate the potential of seed-associated endophytes as plant growth-promoting agents under both controlled and field conditions.
Results
Analysis of Cannabis seed endophyte composition and diversity across 46 plant genotypes revealed 813 different bacterial genera with a predominance of Gammaproteobacteria, Bacilli, Actinobacteria and Alphaproteobacteria but a genotype-specific microbiome. The assessment of domestication and breeding on microbial assembly revealed a higher bacterial diversity in low domestication genotypes (Shannon index, H′: 1.21 vs. 1.05) and a higher homogeneity in bacterial composition caused by line development. Further, a seed bacterial isolate (Bacillus frigoritolerans C1141) associated with low domestication genotypes, and with genes associated with bio-fertilization, bioremediation and phytohormone production, increased plant growth by 42.3% at the time of harvest, under field conditions.
Conclusion
This study addresses critical knowledge gaps related to the assembly of the Cannabis seed-endophytic microbiome. It reveals that Cannabis breeding is linked to alterations of seed microbial communities, which potentially led to the loss of bacteria with functional significance. These results highlight the importance of preserving seed microbiomes in plant breeding to support sustainable plant health and growth enhancement in Cannabis.
Video Abstract – click link below to view
( https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-024-01951-5 )
Background
Plants host microbial communities that are crucial for their growth and vitality [1]. These microbes play multifaceted roles from nutrient mobilization to stress mitigation and enhanced defence against pathogens [2]. Seeds, pivotal in the continuum of plant existence, serve not merely as vessels of plant propagation but as reservoirs of microbial diversity, integral to plant success. Seed-associated microorganisms wield their influence on plants by steering the course of seed germination, preservation, and early development [3]. Furthermore, seed endophytes can be transferred across generations, fostering symbiotic and mutualistic relationships with plants that lay the foundation for robust plant establishment [4,5,6]. Consequently, seeds emerged as promising sources for the isolation of beneficial microorganisms, as well as targets for microbiome-based breeding strategies [7, 8]. However, a comprehensive understanding of seed microbiome structures and functions remains limited, particularly within the Cannabis genus.
Cannabis, a herbaceous annual plant belonging to the Cannabaceae family, boasts a rich history of domestication alongside significant potential for future utilization [9]. Originating in eastern Asia, Cannabis has transcended borders to become a cosmopolitan crop valued for its versatility in providing fibre, oil, food, and bioactive compounds for medicinal as well as recreational purposes. The human-driven selection across diverse regions has contributed to its extensive diversification [10], resulting in predominantly hybrid genotypes today [11]. Notably, the accumulation of tetrahydrocannabinol (THC), which has psychoactive properties, in the trichomes of female flowers, has posed challenges in resource management and genetic enhancement of Cannabis [12]. Recent trends favour cannabidiol (CBD)-rich genotypes for therapeutic purposes, reflecting a projected global market value of US $75.09 billion by 2029 [13]. However, scaling up Cannabis production brings forth challenges, including heightened susceptibility to fungal pathogens and bacterial infections in greenhouse settings and under field conditions [14]. Additionally, mitigating the environmental impact of cultivation practices is essential to align with climate objectives while meeting production demands [15].
Through the processes of plant domestication and breeding, humans have tailored crops to specific quality traits, reshaping the very essence of plant physiology, morphology, and genetic makeup, which has resulted in a discernible reduction in allelic diversity [16]. These processes might have potential repercussions for genes involved in plant-microbiome interactions and thus affect key traits in plant anatomy, immunity or production of secondary metabolites [17]. For instance, alterations in root morphology and exudate profiles of different plant genotypes can alter the selective recruitment of microorganisms from soil [18,19,20,21,22]. Under domestication, seeds often exhibit the most striking phenotypic alterations, emerging as an optimal domain for exploring those effects on microbial communities [23]. Still, little is known about the intricate nuances of these processes on seed microbial assembly.
Previous studies have outlined the differences in microbial communities of different Cannabis genotypes across plant compartments [24,25,26,27]. From those, only a recent study described the variations in bacterial diversity in the Cannabis seed microbiome, however, limited to genotypes with low THC content [28]. Furthermore, the question of whether Cannabis breeding influenced microbial assembly in seeds is left unanswered. Here, we conducted an extensive characterization of the Cannabis bacterial seed microbiome across 46 genotypes with different domestication grades and chemotypes, to cover the high variability of the genus. Seed accessions from early domestication genotypes obtained through natural and artificial selection were included; they are represented by landraces and selected lines. We also included modern genotypes that were obtained through artificial selection; they are represented by accessions classified as cross hybrids and inbred lines. We hypothesized that the heterogeneous genetic background of Cannabis reflects on its seed microbiome. The potential of Cannabis seed endophytes as plant growth-promoting (PGP) and biocontrol agents against different pathogenic fungi has been previously shown [29]. In this work, we tested the potential of Cannabis bacterial endophytes, associated with low domestication genotypes, in promoting growth of Cannabis under controlled and field conditions. We further searched for potential PGP functions in the best-performing bacterium, using whole genome sequencing (WGS). The overall goal of this work is twofold: to comprehensively explore the Cannabis seed microbiome and assess whether this was potentially influenced by domestication, as a strategy for harnessing beneficial seed-endophytic microorganisms that can be used as a sustainable alternative for Cannabis cultivation.
Materials and methods
Metabarcoding analysis
Sample collection
In this study, we comprehensively analysed the bacterial seed microbiome of 46 distinct Cannabis genotypes, sourced from multiple institutions and seed companies across Europe. Seeds were obtained from the Crop Research Institute (CRI) in Prague, Czech Republic (n = 2); the Green House Seed Co. in Amsterdam, Netherlands (n = 2); the Leibniz Institute for Plant Genetics and Crop Plant Research (IPK) in Gatersleben, Germany (n = 27); Hanfama GmbH in Graz, Austria (n = 6); Hanfland GmbH in Hanfthal, Austria (n = 1), Botanical Garden of the University of Graz, Austria (n = 1); the Latvian State Forest Research Institute (LVMI) Silava in Salaspils, Latvia (n = 1); and Sensi Seeds in Amsterdam, Netherlands (n = 6). The sampled genotypes encompassed a spectrum of reproductive types, including dioecious (n = 27), monoecious (n = 9), and sub-dioecious populations (n = 2), as well as varying chemotypes, ranging from low cannabinoid (n = 33) to THC- or CBD-enriched genotypes (n = 9). Furthermore, the samples reflected various grades of domestication, classified as landraces (n = 13), defined for the purpose of this paper as genetically heterogeneous genotypes with minimal domestication [30], selected lines derived from landrace populations (n = 4), a variety of cross hybrids bred from diverse Cannabis genotypes (n = 11), and inbred lines obtained from cross hybrids (n = 5), along with one segregating hybrid and two feminized hybrids obtained by selfing (hybrid S1). Here, we classify seed accessions from early domestication genotypes, i.e. landraces and selected lines, as low domestication genotypes, and modern genotypes, i.e. cross hybrids and inbred lines, as high domestication genotypes. We have included several key Cannabis accessions that have significantly contributed to the development of modern genotypes used for industrial purposes in Europe, the USA and Canada, such as Schurig, Havelländer, Carmagnola and Fibrimon. Comprehensive details regarding the Cannabis genotypes utilized in this study are provided in Table S1.
Amplicon library preparation and sequencing
Seeds underwent a pretreatment process to eliminate non-endophytic microorganisms. Initially, seeds were soaked in sterile deionized water for 4 h on a shaker at 125 rpm. Subsequently, surface sterilization was conducted using a 4% solution of sodium hypochlorite (NaClO) for 5 min with agitation, followed by three 5-min rinses in sterile water. The efficacy of sterilization was confirmed by inoculating 100 μL of the final rinse water onto nutrient agar II (NA II) plates. Seeds were then germinated under sterile conditions until the emergence of radicle and cotyledons. Soaking and germinating seeds can provide a better picture of the endophytic communities by allowing possible rare microorganisms to be activated and multiply to detectable levels [31]. Ten replicate samples, comprising two seedlings each, were obtained for each genotype, except for ID = C86 (8 replicates) and ID = C49 (9 replicates), resulting in a total of 457 samples. This number of replicates allows to sufficiently address the intra-genetic variety of seed-propagated Cannabis genotypes [32]. Subsequently, samples were ground with a pestle in 4 mL of 0.85% NaCl under sterile conditions, and the resulting homogenate was pelleted for further processing. Total DNA extraction was performed using the FastDNA™ SPIN Kit for Soil and the FastPrep Instrument (MP Biomedicals, Santa Ana, CA, USA), following the manufacturer’s instructions. DNA quality was assessed using a NanoDrop 2000 (Thermo Scientific, Wilmington, DE, USA), and samples were stored at − 20 °C for subsequent PCR reactions. The 16S rRNA gene V4 hypervariable region was amplified using the 515f/806r primer pair (515f: 5′-GTGYCAGCMGCCGCGGTAA-3′; 806r: 5′-GGACTACNVGGGTWTCTAAT-3′) [33], with peptide nucleic acid (PNA) clamps employed to block amplification of plastid and mitochondrial 16S rRNA genes during PCR [34]. PCR reactions were performed in a total volume of 30 µL and in three technical replicates using 5 × Taq-&GO Ready Mix (MP Biomedicals, Illkirch, France), 0.2 µM of each primer, 1.5 µM PNA mix (1:1), PCR-grade water, and 3 µL of DNA template. PCR amplification was conducted for 35 cycles after initial denaturation at 96 °C for 5 min, with denaturation at 96 °C for 30 s, PNA annealing at 78 °C for 5 s, primer annealing at 54 °C for 30 s, and elongation at 74 °C for 30 s, followed by a final elongation at 72 °C for 10 min. Amplification products were purified using the Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA) and pooled at equimolar concentrations in three different pools, each including negative controls for DNA extraction and PCR, and PCR products amplified from the ZymoBIOMICS Microbial Community DNA Standard (Zymo Research, Orange, CA, USA) to infer on the bias and errors introduced by sequencing library preparation. Barcoded Illumina libraries were generated and subjected to paired-end sequencing on an Illumina NovaSeq Instrument (Novogene Co., Ltd., UK). The resulting 16S rRNA gene amplicon dataset was deposited in the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena) under the accession number PRJEB64469.
Bioinformatic processing
Initial processing involved quality checking and demultiplexing of pair-end reads using Cutadapt v4.2 [35]. Subsequently, the DADA2 algorithm within QIIME2 v2023.5 was employed for read quality filtering, denoising, read merging, and generation of representative sequences as amplicon sequence variants (ASVs) and the feature table [36, 37]. Taxonomic classification was performed utilizing the VSEARCH algorithm implemented in QIIME2, with the Silva v138 database utilized as the reference for bacterial 16S rRNA gene sequences [38, 39]. Following classification, unassigned and nontarget sequences, including chloroplasts, mitochondria, and archaea, were removed from further analysis. Additionally, samples with fewer than 1000 reads (n = 6) were excluded to ensure a quality-filtered dataset. Contaminant ASVs were identified and eliminated from the dataset using decontam [40] in R v4.2.3 [41] based on prevalence utilizing the fisher method. Ultimately, a total of 36,996,902 high-quality reads, with a mean of 82,033 reads per sample, were retained, resulting in 5297 bacterial ASVs across 451 samples derived from 46 distinct Cannabis genotypes.
Amplicon data analysis
Data manipulation, statistical analysis, and representation were conducted using R v4.2.3 [41] with tidyverse unless otherwise specified. The feature table and taxonomic information were analysed using phyloseq [42]. To ensure comparability, a dataset rarefied to 5607 reads per sample was obtained using rarefy_even_depth from phyloseq on the quality-filtered dataset (Fig. S1). The rarefied dataset was utilized to calculate observed ASV richness and Shannon H′ index. Normality of alpha-diversity measures was assessed using shapiro.test from stats, followed by analysis of variance using kruskal.test, modelling these variables as a function of Cannabis genotype and domestication grade. Pairwise comparisons were calculated using pairwise.wilcox.test with the Bonferroni adjustment method. A heat tree was generated using metacoder [43] down to the genus level with a minimum relative abundance cut-off of 1e-5 for visualization purposes. For beta-diversity calculations, phyloseq_transform_css from metagMisc [44] was employed to normalize counts using cumulative sum scaling (CSS) transform on the quality-filtered dataset. The Bray–Curtis dissimilarity matrix was then calculated using vegdist and subjected to permutational multivariate analysis (PERMANOVA, 999 permutations) with adonis2 in vegan [45] to test differences in microbial communities based on Cannabis genotype, chemotype and domestication grades. Pairwise comparisons were performed using pairwise.adonis in pairwiseAdonis, with false discovery rate correction [46]. Results were projected with UMAP based on Bray–Curtis dissimilarity [47], and silhouette information was extracted from clustering using cluster [48]. The most abundant bacteria (detection > 0.05) were plotted within each genotype with plot_composition from microbiome [49] and the core microbiome across different domestication grades determined with core_members. To further explore the intricate connection between the seed microbiome and Cannabis domestication, we modelled our data with machine learning (ML) using multi-class classification with Gradient Boosted Trees (XGBoost) [50] via tidymodels [51]. Using the trained model, we explained the relative contribution of each ASV to the ML model’s predictions using a feature importance score based on SHAP (SHapley Additive exPlanations) values estimated with shapviz [52]. Features were preprocessed based on a Kruskal–Wallis test (α = 0.05) relative to the domestication grade and abundance data, normalized for training using the CSS transform. To evaluate the predictive performance of the models, we employed five repetitions of tenfold cross-validation and assessed the area under the receiver operating curve (AUC). Finally, we tailored our feature importance score to prioritize abundant taxa. First, we kept only the SHAP values of the overly abundant taxa associated with the domestication level; then, we estimated the average SHAP values of each of the taxa, weighted by the min–max normalized abundance values. Detailed model hyperparameters are provided in Table S2.
Preselection of bacterial strains
Bacterial isolation and identification
Bacterial isolates were obtained from surface-sterilized, germinated seeds of Cannabis sativa L. as detailed above and subsequently cultured on nutrient agar (NA II, SIFIN, Berlin, Germany) amended with nystatin (25 μg/mL). For DNA extraction, bacterial cells underwent thermal lysis at 100 °C for 10 min in a buffer (pH 8) containing 10-mM Tris–Cl, 1 mM EDTA, and 1% Triton X-100, followed by rapid cooling to − 20 °C. DNA integrity was verified using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The full-length 16S rRNA gene was amplified using primers 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492r (5′-TACGGYTACCTTGTTACGACTT-3′). PCR conditions included an initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 61 °C for 30 s, and elongation at 72 °C for 90 s, concluding with a final elongation at 72 °C for 10 min. PCR products were purified with the Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA) and sequenced via Sanger sequencing (LGC Genomics, Berlin, Germany). Sequences were submitted to manual quality filtering with BioEdit 7.7 [53] and analysed using the NCBI BLAST + blastn megablast tool hosted on Galaxy (https://usegalaxy.org/). Only sequences with 100% match to the V4 region of the reference metabarcoding sequences were considered for the identification of the selected bacterial markers.
Bacterial treatments and plant-growing conditions
Selected bacterial strains were cultured individually in Nutrient Broth II (NB II, Difco) at 30 °C overnight until reaching the exponential phase of growth. Cultures were purified through two rounds of centrifugation at 5000 rpm for 10 min, and the resulting bacterial pellets were resuspended in a 0.85% NaCl solution, whereas bacterial concentrations were adjusted to 1 × 108 CFU/mL. Seeds of the Cannabis genotype Fedora 17, an inbred line, were exposed to the different bacterial suspensions for 4 h under agitated conditions at room temperature, followed by germination in sterile CYG germination pouches (MEGA International), each soaked with 15 mL of sterile dH2O. Germinating seeds were kept in dark conditions for 5 days and subsequent 2 days in a greenhouse under a 16-h light cycle at 25 °C. Control seeds were incubated in sterile 0.85% NaCl. The experimental design included three technical replicates per treatment, each comprising 40 seeds. To evaluate the plant growth-promoting effects of the different treatments, data analysis was performed in R v4.2.3 using compare_means from ggpubr [54], applying a nonparametric Wilcoxon rank-sum test with Bonferroni correction for multiple comparisons. The distribution of the variables was assessed using the shapiro.test from stats [41]. For categorical outcomes, the chisq.test with Bonferroni correction for multiple comparisons was employed to analyse the differences between treatments and control.
Field trials
Bacterial treatments and field design
The field trial was conducted at the Experimental Station for Special Cultures (Versuchsstation für Spezialkulturen) in Wies, Austria, within a 187.5 m2 plot divided into three blocks with 1.5 m distance between them. The experimental design included treatment with Bacillus frigoritolerans C1141 (ASV 3055), isolated from the endosphere of Cannabis seeds, and Serratia plymuthica RR2-5–10, isolated from the rhizosphere of oilseed rape cultivated in weakly loamy sand near Rostock, Germany [55]. The latter has been documented for its beneficial effects in various plant systems [56]. Each block consisted of 30 Cannabis plants originated from cuttings of Eletta Campana, a cross hybrid, totalling 90 plants for the experiment. Bacterial stock solutions were prepared from overnight cultures; on the planting day, these cultures were centrifuged, and the cell pellets were resuspended in 0.85% NaCl. These preparations were then diluted to a final concentration of 3 × 107 CFU/mL in the field. The treatment protocol involved immersing the roots of the Cannabis plants in the bacterial suspensions for 5 min before planting, followed by the addition of 50 mL of the suspension to the rhizosphere of each plant. Control plants were treated with the same volume of sterile water. The soil at the Experimental Station for Special Cultures was loamy with a pH of 6.6, 4.4% of organic matter, 18 cmol + /kg calcium, a water retention capacity of 23.7% of its weight and with the following nutrient content: > 200 mg/1000 g of plant-available phosphorus, 526 mg/1000 g of plant-available potassium and 183 mg/100 g of plant-available magnesium. The plot was tilled using Geohobel (Rath) and irrigated before planting. No irrigation, fertilizers, or pesticides were applied during the trial. The concentrations of NO3 and NH4 in the soil were 7 kg/ha and 24 kg/ha, respectively. Recorded climatic conditions from June to September showed average temperatures ranging from 13.7 to 25.9 °C, with total rainfall measuring 155.7 mm. August was the wettest month with 258.4 mm of rain, while September recorded the least precipitation at 49.4 mm. Growth metrics, including plant height and stalk diameter, were collected monthly and final biomass assessments at the study’s conclusion.
Statistical data analysis for plant growth parameters
To evaluate the plant growth-promoting effects, data analysis was performed in R. Differences in growth outcomes between treatments were determined using the compare_means from ggpubr [54], applying a nonparametric Wilcoxon rank-sum test with Bonferroni correction for multiple comparisons. The distribution of the variables was assessed using the shapiro.test from stats [41].
Whole genome sequencing
DNA extraction and PacBio sequencing
DNA from B. frigoritolerans C1141 was extracted from overnight cultures in NB II (OD600 = 2.46) for WGS. The extraction was performed using the MasterPure Complete DNA and RNA Purification Kit (Epicentre, Madison, WI, USA), following the manufacturer’s instructions. Additional steps were incorporated for enhanced lysis efficiency, including supplementation of the lysis buffer with lysozyme and transferring of the samples into a Lysing Matrix E tube containing 1.4-mm ceramic spheres, 0.1-mm silica spheres, and one 4-mm glass sphere for mechanical lysis using a FastPrep-24 instrument (MP Biomedicals) at 6 m/s for 25 s, twice, keeping samples 1 min on ice between repetitions. The concentration of extracted DNA was measured using the Qubit dsDNA HS Assay Kit on a Qubit 4 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). Subsequently, the DNA was subjected to PacBio sequencing on a PacBio Sequel II/IIe platform after SMRTbell library construction (Novogene Co., Ltd., UK).
Genome processing and mining
The continuous long-read sequencing (CLR) single reads obtained (n = 370,273) were employed for de novo assembly using Flye v2.9.1 [57], resulting in a closed circular chromosome spanning 5,620,668 bp. To assess genome completeness and contamination levels, the assembled genome was evaluated using CheckM v1.2.2 [58], estimating values of 98.81% and 1.42%, respectively. Taxonomic classification was verified using GTDB-Tk v1.4.1 [59], and gene annotation was performed using DRAM v1.2.2 [60] resulting in the identification of 5518 predicted protein coding sequences. Subsequently, amino acid sequences generated by DRAM were utilized to annotate bacterial PGP functions on PlaBAse v.1.02 employing strict mode blast + hmmer. The assembled genome, along with the associated annotations, was deposited in the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/) under accession number PRJNA1113337.
Results
Diversity of Cannabis seed endophytes changes with domestication grade
Alpha-diversity analysis, as measured by observed ASV richness and Shannon H′ index, was conducted based on a rarefied dataset (Fig. 1) Across the dataset, observed ASV richness ranged from 10 to 177 ASVs, with a median of 34 (IQR = 57–20.25) and Shannon H′ index from 0.14 to 2.58, with a median of 1.18 (IQR = 1.73–0.59) (Table S3). Significant differences in alpha diversity were found between genotypes and domestication grades for both measures (P ≤ 0.001, Kruskal–Wallis). Notably, Shannon H′ index values revealed a higher number of significant differences between genotypes, indicating uneven species abundances as a differentiating factor (Fig. S2). Regarding domestication, inbred lines exhibited the lowest diversity in both measures, followed by cross hybrids, whereas the highest diversity was observed in plants of low domestication grades, i.e. landraces and selected lines (Table S3). Differences in observed ASV richness between chemotypes were also significant (P ≤ 0.001) (Fig. S2). Differences in alpha diversity were still observed within the same provider (Table S4).
Observed amplicon sequence variants (ASVs) richness and Shannon H′ index were used to assess seed microbiome diversity within and across Cannabis genotypes and domestication grades. Genotypes with THC content > 0.3% are marked with *. Significant differences were found for both observed ASV richness and Shannon H′ index (P ≤ 0.001, Kruskal–Wallis)
Cannabis seed microbiome structure is genotype specific, and line development reduces its dissimilarities
The taxonomic composition of the Cannabis seed microbiome was investigated, revealing 813 different genera in 101 different classes belonging to 38 different phyla. Here, four predominant bacterial classes, Gammaproteobacteria, Bacilli, Actinobacteria and Alphaproteobacteria, collectively represented 99.49% of the reads in the dataset and 65.57% of ASV richness (Fig. 2A). Bacillus was identified as the most abundant genus with 22.09% of the reads, represented by 256 ASVs. This was followed by Pantoea with 54 ASVs covering 21.68% of the respective reads, while Ralstonia accounted for 137 ASVs, representing 21.64% of the reads. Other taxa with relative abundances above 1% were Pseudomonas (10.23%, 222 ASVs), Rhodococcus (5.04%, 65 ASVs), Kosakonia (4.47%, 14 ASVs), Paenibacillus (2.5%, 66 ASVs), Enterobacter (2.3%, 6 ASVs), Methylobacterium-Methylorubrum (1.83%, 44 ASVs), Brevibacillus (1.7%, 30 ASVs), Rathayibacter (1.36%, 51 ASVs) and Sphingomonas (1.07%, 82 ASVs).
The structure of the Cannabis seed microbiome. A The heat tree shows the relative abundance (node colour) and the number of taxa (node size) of the identified seed endophytes on different taxonomic levels. B Dimensionality reduction of the amplicon sequencing variants (ASVs) using UMAP shows bacterial community composition based on a Bray–Curtis dissimilarity matrix, with significant differences between genotypes and domestication grades (P ≤ 0.001). The inset plot shows the separation distances between samples in the different domestication grades. Colours indicate different domestication grades and shape different chemotypes. C Mean relative abundance of the highly abundant (> 5%) members of the microbiome inhabiting seeds of different genotypes on genus level. Different genera are grouped according to bacterial classes using the same colour hue. Blue corresponds to Gammaproteobacteria, red to Bacilli, violet to Actinobacteria, yellow to Alphaproteobacteria, and pink to Bacteroidia, and the remaining genera are displayed in grey as Other. Genotypes with THC content > 0.3% are marked with *
To explore the influence of plant genotype and domestication grade on seed microbiome composition, ordination based on the Bray–Curtis dissimilarity matrix was conducted and visualized in a UMAP plot (Fig. 2B). Analysis of variance using adonis2 revealed significant differences in beta diversity among different genotypes (P ≤ 0.001), explaining 53.6% of the variations in bacterial composition. Pairwise comparisons demonstrated significant differences in bacterial community composition between the majority of the genotypes, with exceptions noted for specific pairs (Fig. S3). Furthermore, domestication explained 9.66% of the variations in bacterial composition (P ≤ 0.001). Differences in bacterial composition between chemotypes were also significant and explained 6.14% of the variations in bacterial composition (P ≤ 0.001). Moreover, significant differences in beta diversity among genotypes, domestication grades, and chemotypes within the same provider were observed (Table S4). Samples from genotypes associated with line development, i.e. selected lines and inbred lines, showed more positive silhouette coefficients, indicating a homogenization of their bacterial communities (Fig. 2b).
Bacterial taxa with a relative abundance of at least 5% in the whole dataset were assessed for each genotype, resulting in the identification of 26 genera across five classes (Fig. 2C). Additionally, these were visualized for each sample across genotypes (Fig. S4). While all five classes were ubiquitous, their relative abundances varied. Gammaproteobacteria exhibited the highest diversity, dominating in 31 genotypes, followed by Bacilli in 13. Notably, certain genera were dominant in more plant genotypes, such as Bacillus and Ralstonia in 12 genotypes each and Pantoea and Pseudomonas in 11 and 5 genotypes, respectively. Actinobacteria predominantly occurred in one genotype (C54), primarily due to the genus Rathayibacter. The same was observed in C37 regarding Alphaproteobacteria, largely due to Methylobacterium-Methylorubrum. The class Bacteroidia did not emerge as dominant in any genotype, with its presence mainly observed in C77. The genotype C47 exhibited the highest percentage of reads outside these dominant classes and genera, albeit accounting for a minor proportion of the dataset.
Identification of bacterial markers linked to plant domestication
We found that the seed core microbiome for each of the domestication grades representing ASVs shared between at least 75% of the samples within the group with a detection level of 0.1%. We found a similar number of core species within each domestication grade, however representing different cumulative relative abundances in each grade, whereas selection lines and inbred lines values reached up to 81.65% and 96.88%, respectively, while for landraces and cross hybrids core species represented 67.61% and 51.8%, respectively (Fig. 3A). In contrast, inbred lines are notably dominated by the presence of one ASV, presenting a relative abundance of 82.05%, i.e. Pantoea agglomerans (ASV 3353). Furthermore, 8 out of 17 core species found across grades, such as Pelomonas (ASV 2062), Ralstonia (ASV 2280 and ASV 2328), Burkholderia sp. (ASV 2389), Pseudomonas sp. (ASV 3198), P.agglomerans (ASV 3353), Enhydrobacter (ASV 3464) and Rhodococcus erythropolis (ASV 4544), were ubiquitous to all grades and collectively constitute a large fraction of the entire bacterial community with a cumulative relative abundance of 63.32% in the whole dataset (Fig. S5). Inbred lines and selected lines have shown to harbour more unique core members, including Pseudomonas (ASV 3136), Kosakonia (ASV 3369) and Enterobacter (ASV 3380) in inbred lines and Bacillus (ASV 2970), Dyella (ASV 2505) and BD7-11 (ASV 3820) in selected lines. Landraces only have Sphingomonas (ASV 1215) as the unique core member, while cross hybrids have no unique core members.
Abundance, taxonomy, and prevalence of core microbiome across domestication grades. A Cumulative relative abundance and richness of core (red) and flexible (violet) amplicon sequence variants (ASVs) and taxonomy of the core microbiome at the ASV level. B Top 10 ASVs (features, ft.) ranked according to their contribution to the gradient-boosted tree classification in the different domestication grades. Higher mean absolute SHAP values indicate higher importance for classification (AUC = 93.6% ± 0.0043%). C Abundance-occupancy curves showing the core (red) and flexible (violet) fractions of the microbiome as well as the identified biomarkers (pink)
The ML-based analysis with XGBoost revealed distinct microbial signatures associated with each domestication grade. ASVs such as Pseudomonas putida (ASV 3151), Bacillus (ASV 3007) and Pseudomonas (ASV 3140) were among the top 10 bacterial genera contributing to classification of landraces (Fig. 3B). Conversely, selection lines exhibited enrichment of taxa such as Rathayibacter (ASV 4090) and Bacillus sp. (ASV 3055). Cross hybrids included taxa such as Bacillus sp. (ASV 2704 and 2970) and Paenibacillus (ASV 4599, 4674, and 4601). Inbred lines had exclusively P. agglomerans (ASV 3353) as their signature, indicating a potential shift in the seed microbiome. These results underscore the potential of using seed microbiome profiles as biomarkers for plant domestication grades and provide insights into potentially beneficial taxa lost during domestication. The abundance and prevalence of the ASVs identified as biomarkers or core members for each domestication grade are presented in Fig. 3C.
Effects of the seed endophyte B. frigoritolerans C1141 on Cannabis growth and fitness
In light of the preceding findings, our investigation delved into the impact of the identified biomarkers from low domestication genotypes on Cannabis growth and fitness under both controlled and field conditions. From the obtained Cannabis seed isolates, a total of five biomarkers, three of landrace genotypes (ASV 3007: Bacillus sp., 3140: Pseudomonas sp., and 3151: P.putida) and two of selected lines (ASV 4090 Rathayibacter sp. and ASV 3055: Bacillus sp.), were identified with a 100% match in the V4 region and selected for further examination (Table S5).
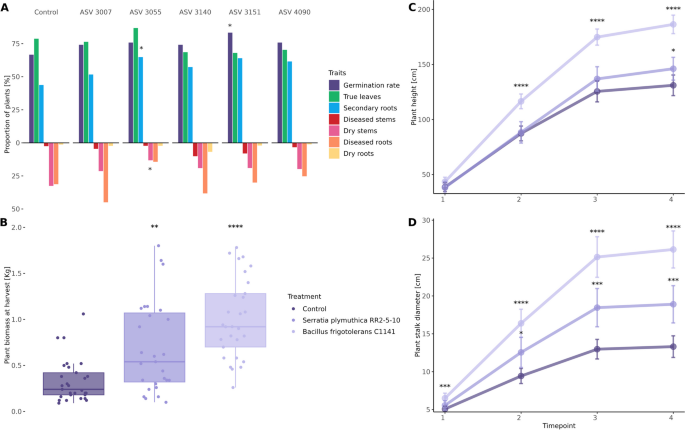
Priming of seeds with the aforementioned bacteria was implemented to assess potential effects on growth under controlled conditions. While all treatments, except for ASV 3140 (Pseudomonas sp.), exhibited enhancements in biomass, root and shoot length in 7-day-old seedlings, these improvements did not reach statistical significance (Fig. S6). Notably, treatment with ASV 3151 (P.putida) significantly increased the germination rate (P ≤ 0.05, χ2) (Fig. 4A), and the treatment with ASV 3055 (Bacillus sp.) emerged as the sole intervention to significantly improve secondary root formation (P ≤ 0.05, χ2) and mitigate the incidence of dead seedlings with dry stems. The latter was further identified as B. frigotolerans, assigned as strain C1141 and selected for further investigation under field conditions.
Influence of the different treatments tested in planta under controlled conditions (A) and in the field (B, C, D). A Comparison between the proportion of observed traits in Cannabis seedlings of the different treatments and the control under controlled conditions. Differences between treated and control Cannabis plants grown under field conditions on their B shoot fresh weight upon harvesting (n = 30 per group) and on their C heights and D stalk diameter at four different time points until the conclusion of the trial, whereas error bars represent the confidence interval of 95%. Significant differences are indicated as follows: * as P ≤ 0.05, ** as P ≤ 0.01, *** as P ≤ 0.001, and **** as P ≤ 0.0001
During field trials, the effects of B. frigoritolerans C1141 on Cannabis plants were compared with those of the patented plant-beneficial bacterium S.plymuthica RR2-5–10, as well as with untreated Cannabis plants. Results showed discernible differences in plant fresh weight upon harvest between the treatment groups and the control, with average yields 3 × higher in plants treated with B. frigoritolerans C1141 and 2 × higher in plants treated with S.plymuthica RR2-5–10 (Fig. 4B). Furthermore, treatment groups exhibited significant differences in plant height over the course of the trial, with marked improvements observed in plants treated with B. frigoritolerans C1141 from the second timepoint on and 43.3% higher at harvest, in contrast to those treated with S.plymuthica RR2-5–10, where an improvement in growth was only evident at the final timepoint (11.5%) (Fig. 4C). Remarkably, plants treated with B. frigoritolerans C1141 demonstrated a significant increase in stalk diameter as early as the first month (timepoint 1), a trend that persisted throughout the duration of the trial, reaching improvements of 96% at the time of harvest, which more than doubles the effects of S.plymuthica RR2-5–10 (42%) (Fig. 4D). These results provided a reliable confirmation of the benefits and adaptability of the seed microbiota when it comes to target approaches for the improvement of plant growth and fitness.
Finally, we conducted a comprehensive analysis of strain C1141’s PGP functions using PLaBase. This approach offers a thorough characterization through genetic analysis, offering detailed insights into the multifaceted mechanisms underlying the potential of strain C1141 as an effective agent for promoting plant growth. A diverse array of 1572 distinct protein coding genes or gene clusters associated with various PGP functions was found and poised positively to impact plant growth, both directly (n = 591) and indirectly (n = 1315) (Fig. 5). Among the notable findings were numerous genes facilitating plant colonization (n = 678), particularly through
the utilization of plant-derived substrates (n = 554) and mechanisms of motility or chemotaxis (n = 60), which are instrumental for seed-endophytic colonization via internal pathways. Additionally, a multitude of genes linked to phytohormone production (n = 254) were identified, with a notable abundance of genes involved in plant vitamin production (n = 132), volatile metabolism (n = 46), and germination stimulation (n = 40). A substantial number of genes associated with bio-fertilization (n = 263) were uncovered, primarily involved in phosphate (n = 120) and potassium (n = 93) solubilization, as well as iron (n = 62) and nitrogen (n = 55) acquisition. Moreover, our analysis revealed genes implicated in immune response stimulation (n = 51), particularly those involved in inducing systemic resistance (ISR) (n = 41), and in stress management or biocontrol (n = 445), aimed at mitigating both biotic (n = 111) and abiotic (n = 306) stress factors. Genes associated with competitive exclusion (n = 490), indicative of bacterial fitness (n = 162), and with mechanisms of quorum sensing or biofilm formation (n = 98) were identified, underscoring their importance in facilitating persistence within the plant ecosystem. Detailed descriptions of the top 25 gene features and associated genes are provided in Table S6.
Predicted PGP functions of B. frigoritolerans C1141. Arches divide the identified PGP functions, represented by the bars, into direct and indirect effects. The height of each bar is shown in log10 scale and describes the number of genes found for each PGP function and the colours group the different PGP functions into different classes
Discussion
In this work, we provide new insights to support harnessing the Cannabis seed microbiome for optimizing crop performance and sustainability in agricultural systems. The plant microbiome plays a pivotal role in shaping plant health and productivity. The process of domestication may have inadvertently disrupted the microbial equilibrium and symbiosis within currently produced plants. This could also include alterations in the microbiome composition and function that are fundamental for optimal growth and resilience against environmental stresses. Given the significant importance of seeds in ensuring the continuity of beneficial microbes, understanding the effects of domestication on the seed microbiome, particularly in economically significant crops like Cannabis, is paramount.
Here, we found evidence that the plant genotype is determinant in explaining the variation in the Cannabis seed bacterial diversity and composition. Although previous research has elucidated the pivotal role of the plant genotype in shaping the assembly of the seed microbiome [61], in Cannabis, it is a novel insight covering a highly heterogeneous genetic background. In analogy to other plant species, the diversity of Cannabis seed microbiome is lower than in other plant compartments [24, 25, 27]. The low moisture content and restricted nutrient availability inherent to seeds create an inhospitable environment for microbial endurance and proliferation [62]. Therefore, seeds are considered a bottleneck in the continuity of the plant microbiome. When compared to other plant species, Cannabis seeds harbour bacterial communities with low to intermediate diversity [63]. Here, differences in evenness of the bacterial communities were more determinant to distinguishing between genotypes than species richness. We have also found well-marked differences between the seed-endophytic community compositions of different Cannabis genotypes. These observations are in accordance with the strong effect of the plant genotype found in other above-ground compartments of Cannabis [24,25,26,27] and are supported by a smaller scale study with 16 genotypes [28]. Similar observations were being made in the seed-endophytic communities of other plant species [64,65,66]. The plant genotype generally shows a higher influence on these microbial communities than geographic location [67,68,69,70]. Although in the present study differences among plant genotypes from the same provider revealed selection by the host, pre- and postharvest variations, such as location, environmental fluctuations or even conditions and duration of storage, can still remain influential [71,72,73]. A detailed untangling of the interplay between host genetics and environmental effects on Cannabis seed microbiome assembly will require targeted experiments.
Domestication was also identified as a significant factor in shaping the seed-endophytic communities of Cannabis. Domestication, and breeding, wielded adeptly, can manipulate the genetic makeup of plants, leading to drastic changes in the seed microbiome, which has also been observed in other plants. Interestingly, the seed community structures differed between domestication grades, and genotypes originating from line development exhibited a higher clustering in comparison to landraces and cross hybrids. Hybridization between different genotypes can introduce novel genetic elements into the plant genome, potentially altering the interactions between the plant and its associated microbiota [74]. On the other hand, inbreeding involves repeated crosses within a limited gene pool, which can lead to the fixation of certain alleles and the loss of others through genetic drift [75]. Similarly, line development involves selecting and propagating individuals with specific traits over multiple generations, often within a narrow genetic background. While these breeding approaches aim to stabilize desired traits, they may inadvertently reduce genetic diversity and the potential for introducing novel genetic elements into the plant genome that could contribute to microbial diversity and interactions. In the present study, a significantly higher bacterial diversity was observed in low domestication grade genotypes (landraces and selected lines) in comparison to high domestication grade (cross hybrids and inbred lines), whereas selected lines showed the highest diversity. Studies have drawn diverging outcomes in this regard [76, 77], which reveal that the effects of domestication on the seed microbiome diversity are dependent on plant species.
In our study, differences between the bacterial composition of low and high THC genotypes were found. In Cannabis, breeding initiatives often coincide with the manipulation of cannabinoid ratios and potency and terpenoid profiles [78]. However, the impact of the interplay between breeding and secondary metabolites on the seed microbiota remains largely unexplored [79]. In a recent study, THC and THC acid were found to exhibit genotoxic effects towards certain bacteria, while other cannabinoids can show cytotoxic or oxidative effects [80]. It is likely that the accumulation of secondary metabolites within the trichomes of Cannabis flowers can act as a deterrent to pests and pathogens and thereby influence the microbial environment surrounding the developing seed.
Here, we used a broad range of Cannabis genotypes to comprehensively explore the bacterial community in Cannabis seed and the potential effects of different chemotypes. From the 101 different bacterial classes found, five — Gammaproteobacteria, Bacilli, Actinobacteria, Alphaproteobacteriaand Bacteroidia — were ubiquitous to all genotypes. Endophytic bacteria of the same phylogeny are commonly found in the seed endosphere of other plants [63]. In Cannabis, these bacteria have been previously reported in other plant compartments. For instance, bacteria belonging to Firmicutes and Proteobacteria — mostly Bacillus and Pseudomonas — were predominantly detected in the flower and leaf endospheres, while Sphingomonas and Methylobacterium have been previously found on their surfaces and Actinobacteria on the root endosphere [24, 26]. This can allude to recruitment from plant to seed via the internal and floral pathways [81]. Furthermore, these classes do not differ from the ones reported in hemp seeds [28]. However, genera like Flavobacterium, Acidovorax, Herminiimonas, Chryseobacterium and Massilia, therein described as most abundant, compose members of the rare taxa in our study, with relative abundances similar or less than 0.01%. Moreover, neither Flavobacterium nor Herminiimonas are highly prevalent among the analysed genotypes in our study. Such differences could have arisen from the harsh sterilization method applied by the authors. A previous study focusing on the surface sterilization effects on endophytes in tea leaves and stems indicated that Bacillus has the highest susceptibility to high concentrations of NaClO [82]. The extent to which these treatments can affect certain types of taxa and yield false negatives can be dependent on their exact localization inside the seed tissues.
The spanning diversity of Cannabis genotypes tested, covering great part of its versatility and the polymorphic amplitude, allowed to refine the identification of shared ASVs. We found that both selected and inbred lines harboured more unique core members in comparison to the other domestication grades. Herein, ASVs shared within the same domestication grade and can point to potentially important M gene-bacteria associations that were affected by the domestication process. M genes, short for microbiome genes, were recently introduced to classify plant genes that are linked to the microbiome [83]. In addition, generally shared ASVs, ubiquitous to all domestication grades, can provide evidence of evolutionary conservation independently of domestication and chemotype. Core genera like Pantoea, Pseudomonas, Ralstonia and Burkholderia seem to be widespread across seeds of different plant species, which may point to important physiological functions in spermatophytes [63]. From the core microbiome of low to high domestication grades, we observed an enrichment of members of the order Enterobacteriales, i.e. Kosakonia and Enterobacter and Pantoea, in the core microbiome of inbred lines and a reduction/depletion of Bacillales, i.e. Staphylococcus and Bacillus. Notably, a large proportion of P. agglomerans (ASV 3353) was found in the core of inbred lines. P. agglomerans has known plant growth promoting [84, 85], as well as antagonistic properties towards phytopathogens [86], and can be vertically transmitted across plant generations [87]. Previous research has consistently detected a higher abundance of Bacteroidetes in the root-associated microbiome of wild relatives, while an enrichment of Actinobacteria and Proteobacteria was observed with modern crops [88, 89]. These observations might convey the influence of soil management practices and are not fully translated to the microbiome of seeds. In seeds, the influence of domestication on the microbial communities has been reported to be dependent on the resulting plant phenotype [90], and an enrichment in Pantoea has been associated with intensive breeding in other species [76, 77, 91].
While it is not clear what consequences this enrichment might have in terms of plant fitness, it would be interesting to address which M genes might be responsible for it.
Based on the diversity assessments performed in this study, we decided to search for particular ASVs that may support growth or fitness of Cannabis plants but were lost during the domestication process, as suggested by Raaijmakers and Kiers [92]. Hence, we consider shifts in the seed microbiome not only a reflection of the domestication state of the plant but also responsible for plant fitness. The present study used rather short 16S rRNA gene fragments to confirm sequence matches between amplicon data and sequences obtained from isolates. Future approaches would profit from full-length 16S rRNA sequencing of amplicons from seed material for more precise identifications.
The field trial results conducted in this study confirmed the capability of the selected isolate B. frigoritolerans C1141 to promote growth in Cannabis cuttings from a cross hybrid. Most notably, this bacterium outperformed the beneficial effects of the already established plant beneficial S. plymuthica RR2-5–10, isolated from the rhizosphere of oilseed rape. These results not only supported seeds, as a useful source of beneficial microorganisms, but also that the modulation of the seed microbiome through personalized interventions may help to restore plant homeostasis and improve plant health. Moreover, these results also apprise to the importance of under-domesticated genotypes like locally adapted landraces, which can provide a repository to reintroduce microbial diversity into domesticated populations. This is especially true in Cannabis whose natural seed microbial transmission got interrupted by phytosanitary requirements or vegetative propagation practices [93]. Furthermore, the comprehensive characterization of B. frigoritolerans C1141 genomic features provided important insights in our understanding of its functional capabilities. This strain harboured a considerable number of genes and gene clusters related to plant colonization, phytohormone production, bio-fertilization, immune response stimulation, stress control and competitive exclusion. These genetic features can explain not only its ability to successfully colonize Cannabis plants and eventually the seeds but also the observed beneficial outcomes in Cannabis.
Conclusion
Our study provides new insights related to the impact of plant genotype, domestication and breeding on endophytic bacterial communities in Cannabis seeds. Cannabis genotypes shared a fraction of predominant bacteria but presented significant differences in terms of diversity and composition. Our data showed further microbial shifts related to domestication and breeding in terms of composition, diversity and abundance. Within the frame of the rewilding hypothesis, we were able to promote Cannabis fitness when reinstating a bacterial seed endophyte associated with low domestication. This work highlights the potential of leveraging seed-associated microorganisms to enhance plant growth and resilience and the importance of considering microbiome-assisted crop improvement or fine-tuning seed endophyte composition via M gene breeding for sustainable Cannabis production by reducing the need for chemical inputs.
Data availability
The assembled Bacillus frigotolerans C1141 genome, along with the associated annotations, was deposited in the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/) under accession number PRJNA1113337. The 16S rRNA gene amplicon dataset was deposited in the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena) under the accession number PRJEB64469. The bioinformatic and statistical analysis pipeline and files for reproducibility are available in the GitHub repository, https://github.com/cbclobato/wild-again.
Abbreviations
- ASVs:
- Amplicon sequencing variants
- AUC:
- Area under the receiver operating curve
- CBD:
- Cannabidiol
- CLR:
- Continuous long-read sequencing
- CSS:
- Cumulative sum scaling
- IQR:
- Interquartile range
- ML:
- Machine learning
- NaClO:
- Sodium hypochlorite
- NA II:
- Nutrient agar II
- NB II:
- Nutrient broth II
- OD600 :
- Optical density at a wavelength of 600 nm
- PGP:
- Plant growth promoting
- PNA:
- Peptide nucleic acid
- SHAP:
- SHapley Additive exPlanations
- THC:
- Tetrahydrocannabinol
- WGS:
- Whole genome sequencing
References
-
Cordovez V, Dini-Andreote F, Carrión VJ, Raaijmakers JM. Ecology and evolution of plant microbiomes. Annu Rev Microbiol. 2019;73(1):69–88. https://doi.org/10.1146/annurev-micro-090817-062524.
-
Trivedi P, Leach JE, Tringe SG, Sa T, Singh BK. Plant–microbiome interactions: from community assembly to plant health. Nat Rev Microbiol. 2020;18(11):607–21. https://doi.org/10.1038/s41579-020-0412-1.
-
Hardoim P. The ecology of seed microbiota. In: Verma SK, White JF Jr, editors. Seed endophytes. Cham: Springer International Publishing; 2019. p. 103–125. https://doi.org/10.1007/978-3-030-10504-4_6.
-
Abdelfattah A, Wisniewski M, Schena L, Tack AJM. Experimental evidence of microbial inheritance in plants and transmission routes from seed to phyllosphere and root. Environ Microbiol. 2021;23(4):2199–214.
-
Shahzad R, Khan AL, Bilal S, Asaf S, Lee IJ. What is there in seeds? Vertically transmitted endophytic resources for sustainable improvement in plant growth. Front Plant Sci. 2018;9:24. https://doi.org/10.3389/fpls.2018.00024.
-
Verma SK, Kharwar RN, White JF. The role of seed-vectored endophytes in seedling development and establishment. Symbiosis. 2019;78(2):107–13. https://doi.org/10.1007/s13199-019-00619-1.
-
Bergna A, Cernava T, Rändler M, Grosch R, Zachow C, Berg G. Tomato seeds preferably transmit plant beneficial endophytes. Phytobiomes J. 2018;2(4):183–93. https://doi.org/10.1094/PBIOMES-06-18-0029-R.
-
Matsumoto H, et al. Bacterial seed endophyte shapes disease resistance in rice. Nat Plants. 2021;7(1):60–72. https://doi.org/10.1038/s41477-020-00826-5.
-
Zhao X, et al. Industrial hemp—an old but versatile bast fiber crop. J Nat Fibers. 2022;19(13):6269–82. https://doi.org/10.1080/15440478.2021.1907834.
-
Clarke RC, Merlin MD. Cannabis domestication, breeding history, present-day genetic diversity, and future prospects. Crit Rev Plant Sci. 2016;35(5–6):293–327. https://doi.org/10.1080/07352689.2016.1267498.
-
Salentijn EMJ, Zhang Q, Amaducci S, Yang M, Trindade LM. New developments in fiber hemp (Cannabis sativa L.) breeding. Ind Crops Prod. 2015;68:32–41. https://doi.org/10.1016/j.indcrop.2014.08.011.
-
Seddon T, Floodgate W. Regulating Cannabis: a global review and future directions. Cham: Springer International Publishing; 2020. https://doi.org/10.1007/978-3-030-52927-7.
-
“Cannabis – worldwide”. Available: https://www.statista.com/outlook/hmo/cannabis/worldwide. Accessed 11 Apr 2024.
-
Punja ZK. Emerging diseases of Cannabis sativa and sustainable management. Pest Manag Sci. 2021;77(9):3857–70. https://doi.org/10.1002/ps.6307.
-
Chakraborty S, Newton AC. Climate change, plant diseases and food security: an overview: climate change and food security. Plant Pathol. 2011;60(1):2–14. https://doi.org/10.1111/j.1365-3059.2010.02411.x.
-
Meyer RS, Purugganan MD. Evolution of crop species: genetics of domestication and diversification. Nat Rev Genet. 2013;14(12):840–52. https://doi.org/10.1038/nrg3605.
-
Gutierrez A, Grillo MA. Effects of domestication on plant–microbiome interactions. Plant Cell Physiol. 2022;63(11):1654–66. https://doi.org/10.1093/pcp/pcac108.
-
Bressan M, et al. Exogenous glucosinolate produced by Arabidopsis thaliana has an impact on microbes in the rhizosphere and plant roots. ISME J. 2009;3(11):1243–57. https://doi.org/10.1038/ismej.2009.68.
-
Cotton TEA, et al. Metabolic regulation of the maize rhizobiome by benzoxazinoids. ISME J. 2019;13(7):1647–58. https://doi.org/10.1038/s41396-019-0375-2.
-
Pérez-Jaramillo JE, et al. Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits. ISME J. 2017;11(10):2244–57. https://doi.org/10.1038/ismej.2017.85.
-
Spor A, et al. Domestication-driven changes in plant traits associated with changes in the assembly of the rhizosphere microbiota in tetraploid wheat. Sci Rep. 2020;10(1):12234. https://doi.org/10.1038/s41598-020-69175-9.
-
Yue H, et al. Plant domestication shapes rhizosphere microbiome assembly and metabolic functions. Microbiome. 2023;11(1):70. https://doi.org/10.1186/s40168-023-01513-1.
-
Soldan R, Fusi M, Cardinale M, Daffonchio D, Preston GM. The effect of plant domestication on host control of the microbiota. Commun Biol. 2021;4(1):936. https://doi.org/10.1038/s42003-021-02467-6.
-
Barnett SE, et al. Evaluating the microbiome of hemp. Phytobiomes J. 2020;4(4):351–63. https://doi.org/10.1094/PBIOMES-06-20-0046-R.
-
Comeau D, Novinscak A, Joly DL, Filion M. Spatio-temporal and cultivar-dependent variations in the Cannabis microbiome. Front Microbiol. 2020;11:491. https://doi.org/10.3389/fmicb.2020.00491.
-
Wei G, et al. Compartment niche shapes the assembly and network of Cannabis sativa-associated microbiome. Front Microbiol. 2021;12:714993. https://doi.org/10.3389/fmicb.2021.714993.
-
Winston ME, et al. Understanding cultivar-specificity and soil determinants of the Cannabis microbiome. PLoS ONE. 2014;9(6):e99641. https://doi.org/10.1371/journal.pone.0099641.
-
Davies J, Hawkins S, Winters A, Farrar K. Bacterial endophytic community composition varies by hemp cultivar in commercially sourced seed. Environ Microbiol Rep. 2024;16(2):e13259. https://doi.org/10.1111/1758-2229.13259.
-
Scott M, Rani M, Samsatly J, Charron J-B, Jabaji S. Endophytes of industrial hemp Cannabis sativa L.) cultivars: identification of culturable bacteria and fungi in leaves, petioles, and seeds. Can J Microbiol. Oct.2018;64(10):664–80. https://doi.org/10.1139/cjm-2018-0108.
-
Villa TCC, Maxted N, Scholten M, Ford-Lloyd B. Defining and identifying crop landraces. Plant Genet Resour. Dec.2005;3(3):373–84. https://doi.org/10.1079/PGR200591.
-
A. Shade et al., “Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity,” mBio, vol. 5, no. 4, pp. e01371–14, Aug. 2014, https://doi.org/10.1128/mBio.01371-14.
-
Z. K. Punja, G. Rodriguez, and S. Chen, “Assessing genetic diversity in Cannabis sativa using molecular approaches”, in Cannabis sativa L. – Botany and Biotechnology, S. Chandra, H. Lata, and M. A. ElSohly, Eds., Cham: Springer International Publishing, 2017, pp. 395–418. https://doi.org/10.1007/978-3-319-54564-6_19.
-
J. G. Caporaso et al., “Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample”, Proc. Natl. Acad. Sci., vol. 108, no. Supplement_1, pp. 4516–4522, Mar. 2011, doi: 10.1073/pnas.1000080107.
-
Lundberg DS, Yourstone S, Mieczkowski P, Jones CD, Dangl JL. Practical innovations for high-throughput amplicon sequencing. Nat Methods. Oct.2013;10(10):999–1002. https://doi.org/10.1038/nmeth.2634.
-
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal. 2011;17(1):10. https://doi.org/10.14806/ej.17.1.200.
-
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–3. https://doi.org/10.1038/nmeth.3869.
-
Bolyen E, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37(8):852–7. https://doi.org/10.1038/s41587-019-0209-9.
-
Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41(D1):D590–6. https://doi.org/10.1093/nar/gks1219.
-
Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. https://doi.org/10.7717/peerj.2584.
-
Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6(1):226. https://doi.org/10.1186/s40168-018-0605-2.
-
R Core Team, R: a language and environment for statistical computing. (2021). R Foundation for Statistical Computing, Vienna, Austria. Available: https://www.R-project.org/
-
McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8(4):e61217. https://doi.org/10.1371/journal.pone.0061217.
-
Foster ZSL, Sharpton TJ, Grünwald NJ. Metacoder: an R package for visualization and manipulation of community taxonomic diversity data. PLOS Comput Biol. 2017;13(2):e1005404. https://doi.org/10.1371/journal.pcbi.1005404.
-
Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. 2013;10(12):1200–2. https://doi.org/10.1038/nmeth.2658.
-
Oksanen J, et al. The vegan package. 2018. Available: https://github.com/vegandevs/vegan.
-
P. Martinez Arbizu, pairwiseAdonis: pairwise multilevel comparison using adonis. (2020). Available: https://github.com/pmartinezarbizu/pairwiseAdonis
-
McInnes L, Healy J, Melville J. UMAP: Uniform Manifold Approximation and Projection for dimension reduction. 2018. https://doi.org/10.48550/ARXIV.1802.03426.
-
Maechler M, Rousseeuw P, Struyf A, Hubert M, Hornik K. cluster: Cluster analysis basics and extensions. 2023. Available: https://CRAN.R-project.org/package=cluster.
-
Lahti L, Shetty S. microbiome R package. 2017. Available: http://microbiome.github.com/microbiome.
-
T. Chen and C. Guestrin, “XGBoost: a scalable tree boosting system,” in Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco California USA: ACM, 2016, pp. 785–794. https://doi.org/10.1145/2939672.2939785.
-
M. Kuhn and H. Wickham, Tidymodels: a collection of packages for modeling and machine learning using tidyverse principles. 2020. [Online]. Available: https://www.tidymodels.org
-
Mayer M, Stando A. SHAP visualizations. 2023. Available: https://github.com/ModelOriented/shapviz.
-
Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Oxf Univ Press. 1999;41:95–8.
-
Kassambara A. ggpubr: ‘ggplot2’ based publication ready plots. 2022. Available: https://cran.r-project.org/web/packages/ggpubr/index.html.
-
Berg G, et al. The rhizosphere effect on bacteria antagonistic towards the pathogenic fungus Verticillium differs depending on plant species and site: rhizosphere-associated antagonistic bacteria. FEMS Microbiol Ecol. 2006;56(2):250–61. https://doi.org/10.1111/j.1574-6941.2005.00025.x.
-
Egamberdieva D, et al. Bacteria able to control foot and root rot and to promote growth of cucumber in salinated soils. Biol Fertil Soils. 2011;47(2):197–205. https://doi.org/10.1007/s00374-010-0523-3.
-
Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37(5):540–6. https://doi.org/10.1038/s41587-019-0072-8.
-
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25(7):1043–55. https://doi.org/10.1101/gr.186072.114.
-
Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2020;36(6):1925–7. https://doi.org/10.1093/bioinformatics/btz848.
-
Shaffer M, et al. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res. 2020;48(16):8883–900. https://doi.org/10.1093/nar/gkaa621.
-
Abdelfattah A, Tack AJM, Lobato C, Wassermann B, Berg G. From seed to seed: the role of microbial inheritance in the assembly of the plant microbiome. Trends Microbiol. 2023;31(4):346–55. https://doi.org/10.1016/j.tim.2022.10.009.
-
War AF, Bashir I, Reshi ZA, Kardol P, Rashid I. Insights into the seed microbiome and its ecological significance in plant life. Microbiol Res. 2023;269:127318. https://doi.org/10.1016/j.micres.2023.127318.
-
Simonin M, et al. Seed microbiota revealed by a large-scale meta-analysis including 50 plant species. Microbiology. 2021. https://doi.org/10.1101/2021.06.08.447541.
-
Bziuk N, et al. The treasure inside barley seeds: microbial diversity and plant beneficial bacteria. Environ Microbiome. 2021;16(1):20. https://doi.org/10.1186/s40793-021-00389-8.
-
Chen X, et al. Nicotiana tabacum seed endophytic communities share a common core structure and genotype-specific signatures in diverging cultivars. Comput Struct Biotechnol J. 2020;18:287–95. https://doi.org/10.1016/j.csbj.2020.01.004.
-
Rybakova D, et al. The structure of the Brassica napus seed microbiome is cultivar-dependent and affects the interactions of symbionts and pathogens. Microbiome. 2017;5(1):104. https://doi.org/10.1186/s40168-017-0310-6.
-
Adam E, Bernhart M, Müller H, Winkler J, Berg G. The Cucurbita pepo seed microbiome: genotype-specific composition and implications for breeding. Plant Soil. 2018;422(1–2):35–49. https://doi.org/10.1007/s11104-016-3113-9.
-
Walitang DI, Kim C, Jeon S, Kang Y, Sa T. Conservation and transmission of seed bacterial endophytes across generations following crossbreeding and repeated inbreeding of rice at different geographic locations. MicrobiologyOpen. 2019;8(3):e00662. https://doi.org/10.1002/mbo3.662.
-
Wassermann B, et al. The Brassica napus seed microbiota is cultivar-specific and transmitted via paternal breeding lines. Microb Biotechnol. 2022;15(9):2379–90. https://doi.org/10.1111/1751-7915.14077.
-
Wolfgang A, et al. Understanding the impact of cultivar, seed origin, and substrate on bacterial diversity of the sugar beet rhizosphere and suppression of soil-borne pathogens. Front Plant Sci. 2020;11:560869. https://doi.org/10.3389/fpls.2020.560869.
-
Chandel A, et al. Implications of seed vault storage strategies for conservation of seed bacterial microbiomes. Front Microbiol. 2021;12:784796. https://doi.org/10.3389/fmicb.2021.784796.
-
Klaedtke S, et al. Terroir is a key driver of seed-associated microbial assemblages: terroir shapes the seed microbiota. Environ Microbiol. 2016;18(6):1792–804. https://doi.org/10.1111/1462-2920.12977.
-
Morales Moreira ZP, Helgason BL, Germida JJ. Crop, genotype, and field environmental conditions shape bacterial and fungal seed epiphytic microbiomes. Can J Microbiol. 2021;67(2):161–73. https://doi.org/10.1139/cjm-2020-0306.
-
Purugganan MD. Evolutionary insights into the nature of plant domestication. Curr Biol. 2019;29(14):R705–14. https://doi.org/10.1016/j.cub.2019.05.053.
-
Ross-Ibarra J, Morrell PL, Gaut BS. Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc Natl Acad Sci. 2007;104(suppl_1):8641–8. https://doi.org/10.1073/pnas.0700643104.
-
Abdullaeva Y, Ambika Manirajan B, Honermeier B, Schnell S, Cardinale M. Domestication affects the composition, diversity, and co-occurrence of the cereal seed microbiota. J Adv Res. 2021;31:75–86. https://doi.org/10.1016/j.jare.2020.12.008.
-
Kim H, Lee KK, Jeon J, Harris WA, Lee YH. Domestication of Oryza species eco-evolutionarily shapes bacterial and fungal communities in rice seed. Microbiome. 2020;8(1):20. https://doi.org/10.1186/s40168-020-00805-0.
-
Welling MT, Shapter T, Rose TJ, Liu L, Stanger R, King GJ. A belated green revolution for Cannabis: virtual genetic resources to fast-track cultivar development. Front Plant Sci. 2016;7:1113. https://doi.org/10.3389/fpls.2016.01113.
-
Wassermann B, Rybakova D, Müller C, Berg G. Harnessing the microbiomes of Brassica vegetables for health issues. Sci Rep. 2017;7(1):17649. https://doi.org/10.1038/s41598-017-17949-z.
-
Harpaz D, Veltman B, Sadeh Y, Marks RS, Bernstein N, Eltzov E. The effect of cannabis toxicity on a model microbiome bacterium epitomized by a panel of bioluminescent E. coli. Chemosphere. 2021;263:128241. https://doi.org/10.1016/j.chemosphere.2020.128241.
-
Shade A, Jacques M-A, Barret M. Ecological patterns of seed microbiome diversity, transmission, and assembly. Curr Opin Microbiol. 2017;37:15–22. https://doi.org/10.1016/j.mib.2017.03.010.
-
Yu Y, Chen Z, Xie H, Feng X, Wang Y, Xu P. Overhauling the effect of surface sterilization on analysis of endophytes in tea plants. Front Plant Sci. 2022;13:849658. https://doi.org/10.3389/fpls.2022.849658.
-
Su P, et al. Microbiome homeostasis on rice leaves is regulated by a precursor molecule of lignin biosynthesis. Nat Commun. 2024;15(1):23. https://doi.org/10.1038/s41467-023-44335-3.
-
Feng Y, Shen D, Song W. Rice endophyte Pantoea agglomerans YS19 promotes host plant growth and affects allocations of host photosynthates. J Appl Microbiol. 2006;100(5):938–45. https://doi.org/10.1111/j.1365-2672.2006.02843.x.
-
Rahman MM, et al. Consistent associations with beneficial bacteria in the seed endosphere of barley (Hordeum vulgare L.). Syst Appl Microbiol. 2018;41(4):386–98. https://doi.org/10.1016/j.syapm.2018.02.003.
-
Links MG, Demeke T, Gräfenhan T, Hill JE, Hemmingsen SM, Dumonceaux TJ. Simultaneous profiling of seed-associated bacteria and fungi reveals antagonistic interactions between microorganisms within a shared epiphytic microbiome on T riticum and B rassica seeds. New Phytol. 2014;202(2):542–53. https://doi.org/10.1111/nph.12693.
-
Ferreira A, Quecine MC, Lacava PT, Oda S, Azevedo JL, AraÃojo WL. Diversity of endophytic bacteria from Eucalyptus species seeds and colonization of seedlings by Pantoea agglomerans. FEMS Microbiol Lett. 2008;287(1):8–14. https://doi.org/10.1111/j.1574-6968.2008.01258.x.
-
Pérez-Jaramillo JE, Carrión VJ, De Hollander M, Raaijmakers JM. The wild side of plant microbiomes. Microbiome. 2018;6(1):143. https://doi.org/10.1186/s40168-018-0519-z.
-
Pérez-Jaramillo JE, Mendes R, Raaijmakers JM. Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol Biol. 2016;90(6):635–44. https://doi.org/10.1007/s11103-015-0337-7.
-
Soldan R, et al. Consistent effects of independent domestication events on the plant microbiota. Curr Biol. 2024;34(3):557-567.e4. https://doi.org/10.1016/j.cub.2023.12.056.
-
Michl K, et al. Determining the footprint of breeding in the seed microbiome of a perennial cereal. Environ Microbiome. 2024;19(1):40. https://doi.org/10.1186/s40793-024-00584-3.
-
Raaijmakers JM, Kiers ET. Rewilding plant microbiomes. Science. 2022;378(6620):599–600. https://doi.org/10.1126/science.abn6350.
-
Berg G, Raaijmakers JM. Saving seed microbiomes. ISME J. 2018;12(5):1167–70. https://doi.org/10.1038/s41396-017-0028-2.
Acknowledgements
The authors would like to thank the staff of the Experimental Station for Special Cultures (Versuchsstation für Spezialkulturen) in Wies, Austria, and particularly Doris Lengauer, for their extensive support during the field trials and for providing the data on soil properties. Furthermore, we would like to thank Katherine Cuadros-Patiño from the Institute of Environmental Biotechnology (TU Graz) for conducting preliminary laboratory trials, Monika Schneider-Trampitsch for helping with the preparation of the field trials, and Matthias Schweitzer, Kristina Michl, Nina Bziuk, Jasper Schierstaedt, Anna Mitterrutzner, Mathias Eisenhut and Rok Pekušek for helping during the field trials. This publication was supported by TU Graz Open Access Publishing Fund.
Funding
Open access funding provided by Graz University of Technology. This study received funding from Austrian Research Promotion Agency (FFG) in the frame of the CannaBiom project (no. 883706) awarded to TC.
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
40168_2024_1951_MOESM1_ESM.docx
Supplementary Material 1: Fig. S1 Alpha rarefaction at 5607 reads per sample represented across (a) genotypes, (b) domestication grades and (c) chemotypes. Fig. S2 Pairwise comparison in (a) ASV-based observed richness and (b) Shannon H’ diversity for the different genotypes. Pairwise comparison in (c) ASV-based observed richness and (d) Shannon H’ diversity for the different domestication grades. Pairwise comparison in (e) ASVbased observed richness and (f) Shannon H’ diversity for the different chemotypes. Significant differences (α = 0.05) are marked (*). Fig. S3 Pairwise comparison of the Bray-Curtis dissimilarity matrix between (a) Cannabis genotypes, (b) domestication grades and (c) chemotypes. Significant differences (α = 0.05) are marked (*). Fig. S4 Bacterial composition at genus level in replicate samples across genotypes (detection level >0.05). Fig. S5 Abundance, taxonomy, and prevalence of core microbiome across the whole dataset represented by ASVs shared between at least 75% of the samples between domestication grades with a detection level of 0.1%. (a) Abundance-occupancy curves showing the core (red) and flexible (violet) fractions of the microbiome. (b) Cumulative relative abundance and richness of core (red) and flexible (violet) amplicon sequencing variants (ASVs) and (c) taxonomy of the core microbiome at the ASV level. Fig. S6 Influence of the different treatments in planta under controlled conditions regarding plant length (a) and biomass (b). Error bars represent the 95% confidence interval. Table S1 Detailed information on the Cannabis genotypes used in this study. LC refers to THC content <0.3%. Table S2 Hyperparameters of gradient boosted-trees. Table S3 Statistics of the calculated alpha diversity indices described by median and interquartile range (IQR) for each Cannabis (a) genotype, (b) domestication grade and (c) chemotype in the rarefied dataset. Table S4 Permutational multivariate analysis (permanova, 999 permutations) with adonis2 in Vegan and Kruskal-Wallis rank sum test on Cannabis genotype, domestication grade and chemotype within the same provider to assess differences in beta and alpha diversity, respectively. The last row shows the differences between providers. Significant differences (α = 0.05) are marked (*). Table S5 Identity and alignment report between identified biomarker ASVs and isolated bacteria from Cannabis seed endosphere. Table S6 Top 25 gene features found in Bacillus frigotolerans C1141 genome, regarding the number of associated genes.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lobato, C., de Freitas, J.M., Habich, D. et al. Wild again: recovery of a beneficial Cannabis seed endophyte from low domestication genotypes. Microbiome 12, 239 (2024). https://doi.org/10.1186/s40168-024-01951-5
- Received
- Accepted
- Published
- DOIhttps://doi.org/10.1186/s40168-024-01951-5